Gene editing is the insertion, deletion or replacement of DNA at a specific site in the genome of an organism or cell. There are the Zinc finger nucleases (ZFNs), transcription-activator like effector nucleases (TALEN), meganucleases and the clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) system have been used for genome editing. CRISPR/Cas9 is simpler, faster, cheaper, and more accurate than other genome editing methods, therefore many scientists who perform genome editing now use CRISPR.
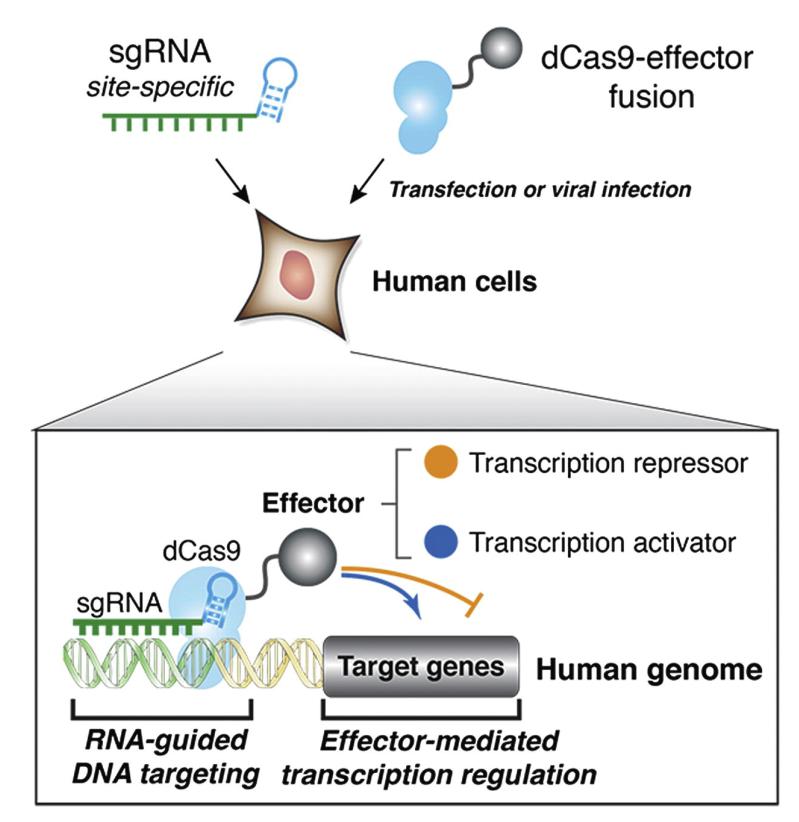
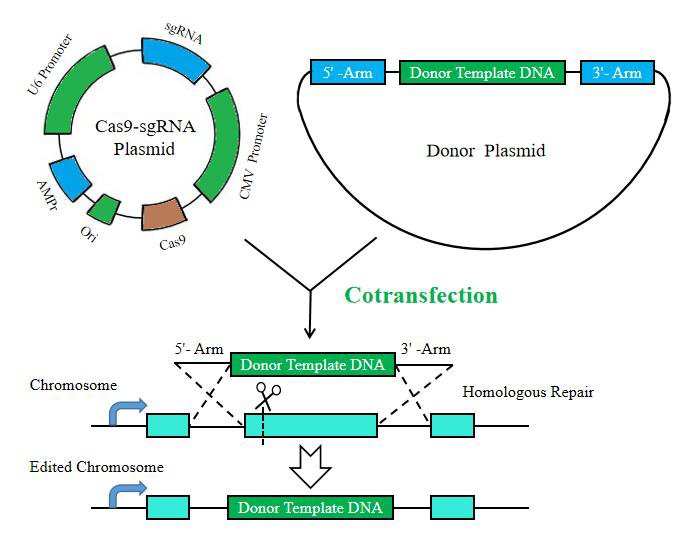
Transcriptional activation Knock-in
BrainVTA is a professional biotechnology company in CRISPR editing field. BrainVTA could offer a range of gene editing services, which is including but not limiting to gene knockout, gene knock-in, transcriptional activation, site-directed mutation. Such genome editing services can offer researchers the comprehensive workflows--from experimental design, to cell line development and validation.
sgRNA screening
|
Screening method |
Advantage |
Disadvantage |
|
Cas9 protein in vitro cleavage assay |
No plasmid required, short cycle |
In vitro screening does not reflect the real state of the genome. |
|
T7 endonuclease I assay |
Closer to the real reaction |
Cell is needed. |
Markers and Reporters
Vectors with mCherry or eGFP reporter genes for monitoring transfection or transduction efficiencies. Stable cell are selected with puromycin marker.
Whole Delivery Formats
● Individual sgRNA or Donor constructs of 5 µg purified plasmid.
● sgRNA screening report.
Key informations
● Fast and high efficiency
● Strong background in CRISPR editing field
● Greater gene-editing efficiency
● Capable of editing multiple genes simultaneously
● The most competitive price
If you have any special requirements in genome editing service, just email us at sales@brainvta.com or click the Send Request button.
Reference:
[1] Eric Murillo-Rodríguez, Rocha N B , André Barciela Veras, et al. The End of Snoring? Application of CRISPR/Cas9 Genome Editing for Sleep Disorders[J]. Sleep and Vigilance, 2018, 2(1):13-21.
[2] Yao, X., Wang, X., Hu, X., Liu, Z., Liu, J., Zhou, H., Shen, X., Wei, Y., Huang, Z., Ying, W., et al. (2017). Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell research 27, 801-814.
[3] Wiles M V , Qin W , Cheng A W , et al. CRISPR–Cas9-mediated genome editing and guide RNA design[J]. Mammalian Genome, 2015, 26(9-10).
[4] Wang H , Yang H , Shivalila C , et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering[J]. Cell, 2013, 153(4):910-918.

